mplpub¶
mplpub is a simple python module that provides a number of templates for publication quality matplotlib figures. It can be installed using pip as follows:
python3 -m pip mplpub
For maximum plot quality and flexibility when including mathematical equations, mplpub uses your system’s LaTeX distribution to render labels etc. Thus you will need a working LaTeX distribution, and the helvet LaTeX package which is used to render all text and math using Helvetica. This package in typically shipped with comprehensive LaTeX distributions, e.g., TeXLive.
Templates¶
Currently the following templates are available:
acs
acs_dcol
base
base_2.0_update
jpcl_toc
natcom
natcom_dcol
phd
phd_fw
rsc
rsc_dcol
Upper-case greek letters¶
Upper-case greek letters are generally not available when using sans-family fonts such as Helvetica.
In this case, one can often get acceptable results by falling back to a serif font.
In mplpub this has been conveniently implemented as a LaTeX command called \UG that can be invoked in you LaTeX-containing python strings like so:
r"Upper-case greek letters: $\UG{\Delta}$"
Commands¶
- mplpub.setup(tex=True, template=None, width=None, height=None, font_family='sans-serif', color_cycle=odict_values(['#1F77B4', '#FF7F0E', '#2CA02C', '#D62728', '#9467BD', '#8C564B', '#E377C2', '#7F7F7F', '#BCBD22', '#17BECF', '#AEC7E8', '#FFBB78', '#98DF8A', '#FF9896', '#C5B0D5', '#C49C94', '#F7B6D2', '#C7C7C7', '#DBDB8D', '#9EDAE5']), extra_settings=None)[source]¶
Set up publication quality plotting by changing the
rcparamsdictionary and various other options.
Examples¶
A simple plot showcasing the basic features of the mplpub plotter.
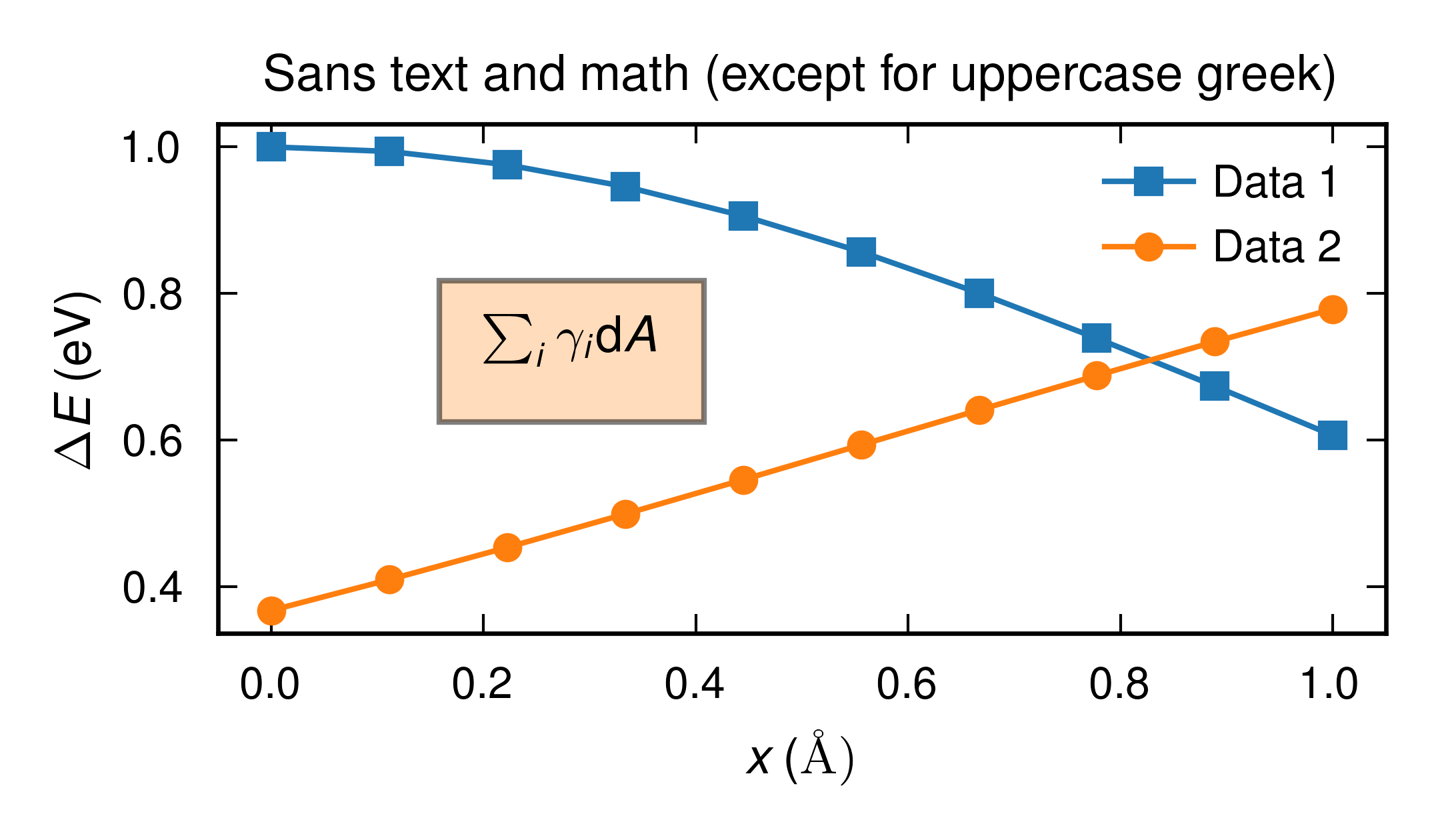
Source code:
import numpy as np import matplotlib.pyplot as plt import mplpub # setup for publication quality plotting using default options, # plot labels etc. will be rendedred by system LaTeX using a Helvetica # sans-serif font mplpub.setup(height=2.1, width=3.6) # plot something x = np.linspace(0, 1, 10) y1 = np.exp(-x**2/2.0) y2 = np.exp(-(x-2)**2/4.0) fig, ax = plt.subplots() ax.plot(x, y1, marker='s', label='Data 1') ax.plot(x, y2, marker='o', label='Data 2') # generally for latex we need an r before the string to not # interpret backslash as an escape sequence # also note that mplpub automatically loads amsmath so we can # use the \text command ax.set_xlabel(r'$x \,\text{(\normalfont\AA)}$') # Glyphs for upper case greek letters are typically not included in sans fonts # so to get them the mplpub module employs a dirty hack where we temporarily # fall back to a serif font, this is handled automatically # by the custom UG command: ax.set_ylabel(r'$\UG{\Delta}E \,(\text{eV})$') # the module comes with nice colors saved in dictionaries, the default # colors are taken from the tableau palette but can also be accessed # through a dictionary ax.text(0.2, 0.7, r'$\sum_i\gamma_i\mathrm{d}A$', bbox={'facecolor': mplpub.tableau['lightorange'], 'alpha': 0.5, 'pad': 8}) ax.set_title(r'Sans text and math (except for uppercase greek)') ax.legend() # legends are automatically located at the "best" position plt.tight_layout() plt.savefig('example_basic.png')
A plot with different x scales (and labels) at the top and bottom.
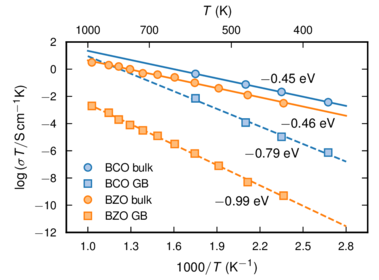
Source code:
import numpy as np import matplotlib.pyplot as plt import mplpub ################################################################### # Data ################################################################### # BaCeO3 data - Bassano et. al., Solid State Ionics 180, 168 (2009): # fig 8, 1250 C-data T_inv_bco = np.array([1.75, 2.1, 2.35, 2.675]) # 1000/T in Kelvin T_bco = 1000.0 / T_inv_bco # T in Kelvin log_s_b_bco = np.array([-3.1, -3.8, -4.3, -5.0]) # log(sigma_bulk) log_s_gb_bco = np.array([-4.9, -6.6, -7.6, -8.7]) # log(sigma_gb) log_sT_b_bco = np.log10(10**(log_s_b_bco) * T_bco) # log(sigma_bulk * T) log_sT_gb_bco = np.log10(10**(log_s_gb_bco) * T_bco) # log(sigma_gb * T) # BaZrO3 data - Shirpour et. al., PCCP 14, 730 (2012): Fig 9, 6YBZ-data T_bzo = 273 + np.array([150, 200, 250, 300, 350, 400, 450, 500, 550, 600, 700]) # T in Kelvin T_inv_bzo = 1000.0 / T_bzo # 1000/T in Kelvin log_sT_b_bzo = np.array([-2.5, -1.9, -1.4, -1.0, -0.6, -0.4, -0.3, 0.0, 0.2, 0.3, 0.5]) # log(sigma_bulk * T) log_sT_gb_bzo = np.array([-9.3, -8.3, -7.1, -6.2, -5.5, -4.9, -4.5, -4.1, -3.7, -3.2, -2.7]) # log(sigma_gb * T) # Fits to data points b_fit_bco = np.polyfit(T_inv_bco, log_sT_b_bco, 1) gb_fit_bco = np.polyfit(T_inv_bco, log_sT_gb_bco, 1) b_fit_bzo = np.polyfit(T_inv_bzo, log_sT_b_bzo, 1) gb_fit_bzo = np.polyfit(T_inv_bzo, log_sT_gb_bzo, 1) temp_inv = np.linspace(1, 2.8, 181) line_b_bco = b_fit_bco[0] * temp_inv + b_fit_bco[1] line_gb_bco = gb_fit_bco[0] * temp_inv + gb_fit_bco[1] line_b_bzo = b_fit_bzo[0] * temp_inv + b_fit_bzo[1] line_gb_bzo = gb_fit_bzo[0] * temp_inv + gb_fit_bzo[1] ################################################################### # Plot settings ################################################################### # Setup for publication quality plotting using ACS (American Chemical # Society) template with a non-default option for figure height mplpub.setup(template='acs', height=2.4) # X and Y label strings x_label_string1 = r'$1000/T$ ($\text{K}^{-1}$)' x_label_string2 = r'$T$ (K)' y_label_string = r'$\log{(\sigma T / \text{S}\,\text{cm}^{-1}\text{K})}$' # X and Y tick postions xmin = 1.0 xmax = 2.8 dx = 0.3 ddx = 0.15 nx = int(round((xmax - xmin) / dx) + 1) x_tick_values1 = xmin + dx * np.array(range(nx)) x_tick_values2 = np.array([400, 500, 700, 1000]) x_tick_positions2 = 1000.0 / x_tick_values2 ymin = -12 ymax = 2 dy = 2 ddy = 0 ny = int(round((ymax - ymin) / dy) + 1) y_tick_values = ymin + dy * np.array(range(ny)) ################################################################### # Plot figure ################################################################### fig, ax1 = plt.subplots() ax1.plot(temp_inv, line_b_bco, '-', color=mplpub.tableau['blue']) ax1.plot(temp_inv, line_gb_bco, '--', color=mplpub.tableau['blue']) ax1.plot(temp_inv, line_b_bzo, '-', color=mplpub.tableau['orange']) ax1.plot(temp_inv, line_gb_bzo, '--', color=mplpub.tableau['orange']) ax1.plot( T_inv_bco, log_sT_b_bco, 'o', markeredgecolor=mplpub.tableau['blue'], markerfacecolor=mplpub.tableau['lightblue'], label=r'BCO bulk') ax1.plot( T_inv_bco, log_sT_gb_bco, 's', markeredgecolor=mplpub.tableau['blue'], markerfacecolor=mplpub.tableau['lightblue'], label=r'BCO GB') ax1.plot( T_inv_bzo, log_sT_b_bzo, 'o', markeredgecolor=mplpub.tableau['orange'], markerfacecolor=mplpub.tableau['lightorange'], label=r'BZO bulk') ax1.plot( T_inv_bzo, log_sT_gb_bzo, 's', markeredgecolor=mplpub.tableau['orange'], markerfacecolor=mplpub.tableau['lightorange'], label=r'BZO GB') # Sets the x- and y-scales ax1.set_xlim([xmin - ddx, xmax + ddx]) ax1.set_ylim([ymin - ddy, ymax + ddy]) ax1.set_xlabel(x_label_string1) ax1.set_ylabel(y_label_string) ax1.set_xticks(x_tick_values1) ax1.set_yticks(y_tick_values) # Sets the top x-scale ax2 = ax1.twiny() ax1.xaxis.set_ticks_position('bottom') ax2.xaxis.set_ticks_position('top') ax2.axes.set_xticks(x_tick_positions2) ax2.axes.set_xticklabels(x_tick_values2, position=(0, 0.98)) ax2.set_xlim([xmin - ddx, xmax + ddx]) ax2.set_xlabel(x_label_string2, labelpad=6.4) ax1.legend() # legends are automatically located at the "best" position ax1.text(2.20, -01.0, r'$-$0.45 eV', fontsize=8) ax1.text(2.34, -04.3, r'$-$0.46 eV', fontsize=8) ax1.text(2.09, -06.5, r'$-$0.79 eV', fontsize=8) ax1.text(1.88, -10.0, r'$-$0.99 eV', fontsize=8) # Adjust figure to remove white space at edges. Can be used even without # subplots. plt.subplots_adjust(left=0.175, bottom=0.17, right=0.97, top=0.85) plt.savefig('example_multiple_xscales.pdf')
A schematic plot with no ticks or quantitative data.
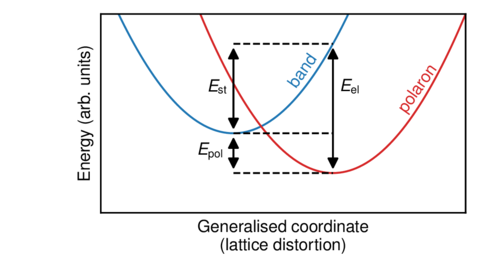
Source code:
import numpy as np import matplotlib.pyplot as plt import mplpub # Schematic illustration without ticks or other quantitative data # Contributed by Erik Jedvik mplpub.setup() x = np.arange(0.0, 1.2, 0.01) band = 5*(x - 0.4)**2 + 0.4 pol = 5*(x - 0.7)**2 + 0.2 fig, ax = plt.subplots() ax.plot(x, band) ax.plot(x, pol, color=mplpub.tableau['red']) ax.plot([0.4, 0.7], [0.2, 0.2], '--k') ax.plot([0.4, 0.7], [0.4, 0.4], '--k') ax.plot([0.4, 0.7], [0.85, 0.85], '--k') ax.set_xlabel('Generalised coordinate\n(lattice distortion)') ax.set_ylabel('Energy (arb. units)') ax.set_xlim([0.0, 1.1]) ax.set_ylim([0.0, 1.0]) # remove ticks and tick labels, the tick_params method will # provide you with the most control ax.tick_params(axis='both', # which axex to operate on which='both', # operate on both major and minor ticks bottom=False, # turn bottom ticks off top=False, left=False, right=False, labelbottom=False, # turns bottom ticklabels off labelleft=False) # an quicker way with little room for customization # ax.set_xticks([]) # ax.set_yticks([]) # now we annotate the schematic figure with some helpful text ax.text(0.56, 0.75, 'band', rotation=55, color=mplpub.tableau['blue']) ax.text(0.89, 0.70, 'polaron', rotation=55, color=mplpub.tableau['red']) ax.text(0.29, 0.28, r'$E_\text{pol}$') ax.text(0.32, 0.6, r'$E_\text{st}$') ax.text(0.72, 0.6, r'$E_\text{el}$') ax.annotate('', xy=(0.4, 0.2), xycoords='data', xytext=(0.4, 0.4), textcoords='data', size=12, va='center', ha='center', arrowprops=dict(arrowstyle='<|-|>', connectionstyle='arc3, rad=0', fc='k') ) ax.annotate('', xy=(0.4, 0.4), xycoords='data', xytext=(0.4, 0.85), textcoords='data', size=12, va='center', ha='center', arrowprops=dict(arrowstyle='<|-|>', connectionstyle='arc3, rad=0', fc='k') ) ax.annotate('', xy=(0.7, 0.2), xycoords='data', xytext=(0.7, 0.85), textcoords='data', size=12, va='center', ha='center', arrowprops=dict(arrowstyle='<|-|>', connectionstyle='arc3, rad=0', fc='k') ) plt.subplots_adjust(left=0.21, bottom=0.24, right=0.97, top=0.95, wspace=0.15, hspace=0.10) plt.savefig('example_schematic.pdf')
A fancy schematic figure that includes png insets (from OVITO in this case).
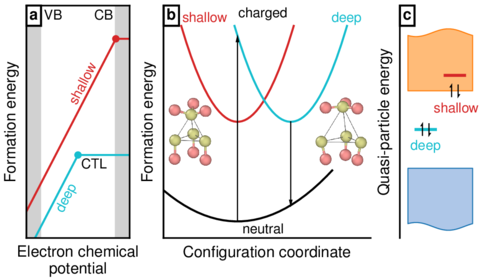
Source code:
#!/usr/bin/env python import numpy as np import matplotlib.pyplot as plt import matplotlib.gridspec as gridspec from matplotlib.offsetbox import OffsetImage, AnnotationBbox from matplotlib._png import read_png import mplpub # Set figure specs mplpub.setup(template='natcom', height=2.0) fig = plt.figure() gs = gridspec.GridSpec(1, 100) ax1 = plt.subplot(gs[0, 2:24]) ax2 = plt.subplot(gs[0, 31:75]) ax3 = plt.subplot(gs[0, 81:100]) fig.subplots_adjust(left=0.035, bottom=0.15, right=1.01, top=0.98) ################################################################### # sub figure 1 ################################################################### # Data to plot mue = np.linspace(0, 3, 31) s = 0.8 + 2 * mue d = -0.5 + 2 * mue ctl_s = 26 ctl_d = 15 s[ctl_s + 1:] = s[ctl_s] d[ctl_d + 1:] = d[ctl_d] be = 0.4 # plot settings xmin = -0 xmax = 3 ymin = 0 ymax = 7 # plot data ax1.plot(mue, d, '-', color=mplpub.tableau['turquoise']) ax1.plot(mue, s, '-', color=mplpub.tableau['red']) ax1.plot( mue[ctl_d], d[ctl_d], 'o', markersize=3, markerfacecolor=mplpub.tableau['turquoise'], markeredgecolor=mplpub.tableau['turquoise'], ) ax1.plot( mue[ctl_s], s[ctl_s], 'o', markersize=3, markerfacecolor=mplpub.tableau['red'], markeredgecolor=mplpub.tableau['red'], ) # grey shaded areas ax1.fill_betweenx( [ymin, ymax], xmin, xmin + be, edgecolor=mplpub.tableau['lightgrey'], facecolor=mplpub.tableau['lightgrey'], ) ax1.fill_betweenx( [ymin, ymax], xmax - be, xmax, edgecolor=mplpub.tableau['lightgrey'], facecolor=mplpub.tableau['lightgrey'], ) # set x- and y-limits ax1.set_xlim([xmin, xmax]) ax1.set_ylim([ymin, ymax]) # remove x- and y-ticks ax1.set_xticks([]) ax1.set_yticks([]) # set axis labels ax1.set_xlabel('Electron chemical\n potential') ax1.set_ylabel('Formation energy') # text pieces. the transform argument yields fractional coordinates # for the text positions, which is very convenient. ax1.text(0.17, 0.94, 'VB', transform=ax1.transAxes, fontsize=7) ax1.text(0.84, 0.94, 'CB', transform=ax1.transAxes, fontsize=7, ha='right') ax1.text( 0.40, 0.72, 'shallow', transform=ax1.transAxes, fontsize=7, rotation=63, color=mplpub.tableau['red'], ) ax1.text( 0.27, 0.20, 'deep', transform=ax1.transAxes, fontsize=7, rotation=63, color=mplpub.tableau['turquoise'], ) ax1.text(0.53, 0.30, 'CTL', transform=ax1.transAxes, fontsize=7) ax1.text( 0.03, 0.94, r'\textbf{a}', transform=ax1.transAxes, bbox={'facecolor': 'white', 'alpha': 1, 'pad': 1.5, 'linewidth': 1}, ) ################################################################### # sub figure 2 ################################################################### # Data to plot y_d_n_x0 = 2.5 y_d_n_y0 = 0.5 y_d_c_x0 = 4.3 y_d_c_y0 = 3.5 nbr_gridpoints = 701 x_range = np.linspace(0, 7, nbr_gridpoints) y_d_n = y_d_n_y0 + 0.15 * (x_range - y_d_n_x0) ** 2 y_d_c = y_d_c_y0 + 0.80 * (x_range - y_d_c_x0) ** 2 y_s_c = y_d_c_y0 + 0.80 * (x_range - y_d_n_x0) ** 2 # plot settings xmin = 0 xmax = 7 ymin = 0 ymax = 7 # plot data ax2.plot(x_range[60:441], y_s_c[60:441], '-', color=mplpub.tableau['red']) ax2.plot(x_range[0:571], y_d_n[0:571], '-k') ax2.plot(x_range[240:621], y_d_c[240:621], '-', color=mplpub.tableau['turquoise']) # set x- and y-limits ax2.set_xlim([xmin, xmax]) ax2.set_ylim([ymin, ymax]) # remove x- and y-ticks ax2.set_xticks([]) ax2.set_yticks([]) # set axis labels ax2.set_xlabel('Configuration coordinate') ax2.set_ylabel('Formation energy') # remove borders ax2.spines['right'].set_visible(False) ax2.spines['top'].set_visible(False) # text pieces. the transform argument yields fractional coordinates # for the text positions, which is very convenient. ax2.text(0.48, 0.02, 'neutral', transform=ax2.transAxes, fontsize=7, ha='center') ax2.text(0.48, 0.95, 'charged', transform=ax2.transAxes, fontsize=7, ha='center') ax2.text( 0.09, 0.93, 'shallow', transform=ax2.transAxes, fontsize=7, color=mplpub.tableau['red'], ) ax2.text( 0.81, 0.93, 'deep', transform=ax2.transAxes, fontsize=7, color=mplpub.tableau['turquoise'], ) ax2.text( 0.015, 0.94, r'\textbf{b}', transform=ax2.transAxes, bbox={'facecolor': 'white', 'alpha': 1, 'pad': 1.5, 'linewidth': 1}, ) # arrows arrow_head_width = 0.08 arrow_head_length = 0.15 ax2.arrow( y_d_c_x0, y_d_c_y0, 0, y_d_n[431] - y_d_c_y0, head_width=arrow_head_width, head_length=arrow_head_length, length_includes_head=True, fc='k', ec='k', zorder=20, linewidth=0.5, ) ax2.arrow( y_d_n_x0, y_d_n_y0, 0, y_d_c[251] - y_d_n_y0, head_width=arrow_head_width, head_length=arrow_head_length, length_includes_head=True, fc='k', ec='k', zorder=20, linewidth=0.5, ) # embedd png images arr_neutral = read_png('example_fancy_schematic_embedded_png_fig1.png') imagebox = OffsetImage(arr_neutral, zoom=0.105) ab = AnnotationBbox(imagebox, [0.9, 3.2], frameon=False) ax2.add_artist(ab) arr_neutral = read_png('example_fancy_schematic_embedded_png_fig2.png') imagebox = OffsetImage(arr_neutral, zoom=0.105) ab = AnnotationBbox(imagebox, [6.0, 3.2], frameon=False) ax2.add_artist(ab) ################################################################### # sub figure 3 ################################################################### # Data to plot ctl_d = [0.75, 1.35] ctl_s = [1.65, 2.25] xstart = 0.5 xend = 2.5 nbr_gridpoints = 1001 x_range = np.linspace(xstart, xend, nbr_gridpoints) y_min_range = 0.5 - 0.1 * np.sin(np.pi / 4.0 + 2 * np.pi * x_range / (xstart - xend)) y_max_range = 5.5 + 0.1 * np.sin(np.pi / 4.0 + 2 * np.pi * x_range / (xstart - xend)) # plot settings xmin = 0.25 xmax = 3.0 ymin = 0.2 ymax = 6.2 # plot valence (blue) and conduction (orange) bands as filled regions ax3.fill_between( x_range, y_min_range, 2.0, edgecolor=mplpub.tableau['blue'], facecolor=mplpub.tableau['lightblue'], linewidth=0.5, ) ax3.fill_between( x_range, 4.0, y_max_range, edgecolor=mplpub.tableau['orange'], facecolor=mplpub.tableau['lightorange'], linewidth=0.5, ) # plot data ax3.plot(ctl_d, [3.0, 3.0], '-', color=mplpub.tableau['turquoise'], linewidth=1.5) ax3.plot(ctl_s, [4.4, 4.4], '-', color=mplpub.tableau['red'], linewidth=1.5) # set x- and y-limits ax3.set_xlim([xmin, xmax]) ax3.set_ylim([ymin, ymax]) # remove x- and y-ticks ax3.set_xticks([]) ax3.set_yticks([]) # set axis labels ax3.set_ylabel('Quasi-particle energy') # remove borders ax3.spines['right'].set_visible(False) ax3.spines['top'].set_visible(False) ax3.spines['bottom'].set_visible(False) # arrows (electrons in defect levels) arrow_head_width = 0.08 arrow_head_length = 0.05 ax3.arrow( 0.95, 2.85, 0, 0.3, head_width=arrow_head_width, head_length=arrow_head_length, length_includes_head=True, fc='k', ec='k', zorder=20, linewidth=0.5, shape='right', ) ax3.arrow( 1.15, 3.15, 0, -0.3, head_width=arrow_head_width, head_length=arrow_head_length, length_includes_head=True, fc='k', ec='k', zorder=20, linewidth=0.5, shape='right', ) ax3.arrow( 1.85, 3.85, 0, 0.3, head_width=arrow_head_width, head_length=arrow_head_length, length_includes_head=True, fc='k', ec='k', zorder=20, linewidth=0.5, shape='right', ) ax3.arrow( 2.05, 4.15, 0, -0.3, head_width=arrow_head_width, head_length=arrow_head_length, length_includes_head=True, fc='k', ec='k', zorder=20, linewidth=0.5, shape='right', ) # text pieces. the transform argument yields fractional coordinates # for the text positions, which is very convenient. ax3.text( 0.380, 0.54, 'shallow', transform=ax3.transAxes, fontsize=7, color=mplpub.tableau['red'], ) ax3.text( 0.120, 0.38, 'deep', transform=ax3.transAxes, fontsize=7, color=mplpub.tableau['turquoise'], ) ax3.text( 0.035, 0.94, r'\textbf{c}', transform=ax3.transAxes, bbox={'facecolor': 'white', 'alpha': 1, 'pad': 1.5, 'linewidth': 1}, ) ################################################################### # save figure to file ################################################################### fig.savefig('example_fancy_schematic_embedded_png.pdf')